Welcome! TSS Analyzer is a sequence-based inference framework for analyzing TSS motifs, aimed to provide an user-friendly interface of single/multiple sequence motif visualizations. The web server is developed by Chenlai Shi. Puffin & Puffin-D are developed by Kseniia Dudnyk and Jian Zhou. Puffin is completely free for any non-commercial and academic use, and please contact us for commercial applications.
In the home page, you can select the prediction mode and provide the corresponding input information. Here we list the different prediction modes that we currently support and the required input information:
Genomic Region - Predict TSS motifs centered at the specified coordinate (for example,
Genome Human (Homo sapiens/hg38), Chromosome chr7, Center Coordinate
5530600, and strand -) from sequence and compare with experimental observations
Gene Name - Predict the sequence specified by gene name (for example, type in Gene Name
ACTB), which also auto-fills the corresponding genomic region. You may also limit your search
among highly expressed tss or search by Gene ID for other mammal species
Sequence - Predict the provided sequence contents. Either edit the sequence populated by the above two approaches or paste in your own sequence
The predictions are visualized with plots of motif activation, motif effects, sum of motif effects, prediction and experimental observations. Motifs are highlighted at corresponding positions of sequence. More from the control panel:
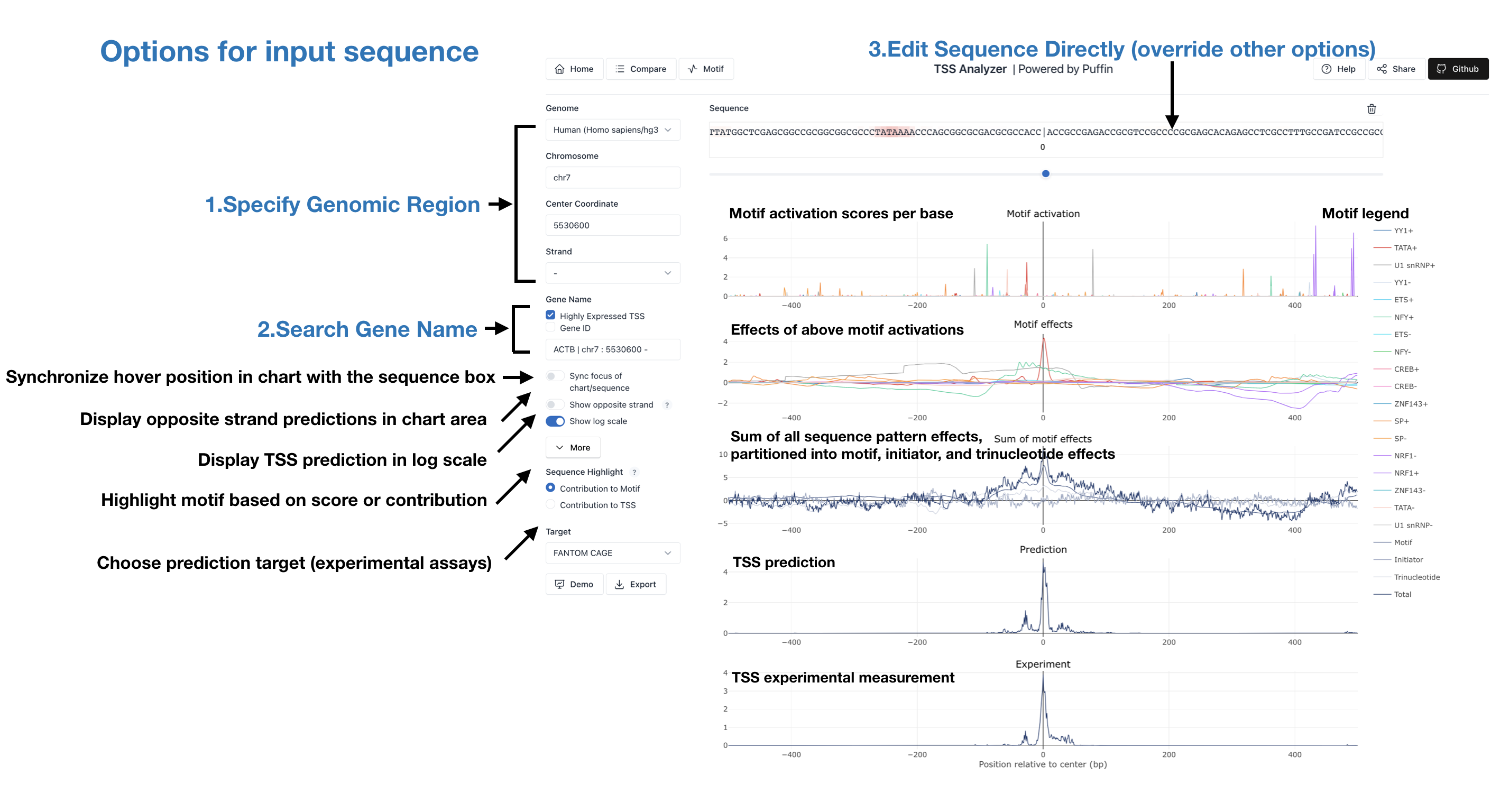
Sync focus of chart/sequence - Synchronize hover position in chart area with corresponding sequence
Show opposite strand - Display the opposite strand predictions in chart area
Show log scale - Display predicted values in log scale
Sequence Highlight - Highlight motif based on Motif contribution or TSS contribution
Target - Choose among different prediction target files
Demo - Show auto-populated examples as demonstration
Export - Export data to files
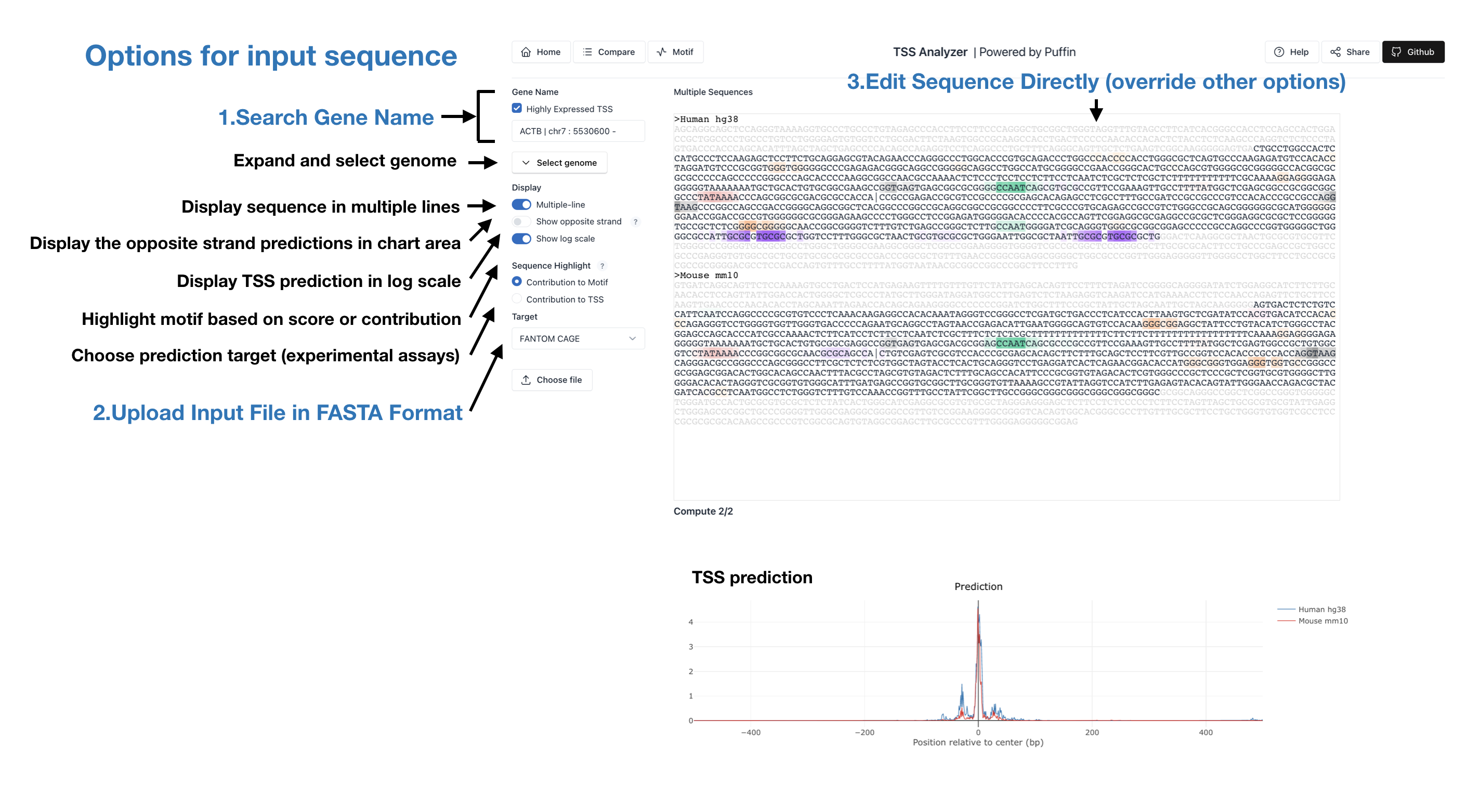
Similar to the setups of home page, you can select the prediction mode and provide the corresponding input information. Here we list the different prediction modes that we currently support and the required input information:
Gene Name - Predict the sequence specified by gene name (for example, type in Gene Name
ACTB). You may also limit your search among highly expressed tss or search by Gene ID for other
mammal species
Choose file - Predict the provided sequence from text file in fasta
format uploaded. Sequences should be separated by line break\n or
>label\n
Sequence - Predict the provided sequence contents. Either edit the sequence populated by
the above two approaches or paste in your own sequence. Sequences should be separated by line
break\n or >label\n
The predictions are visualized in one plot. Motifs are highlighted at corresponding positions of sequence. More from the control panel:
Multiple-line - Display sequence in multiple lines
Show opposite strand - Display the opposite strand predictions in chart area
Show log scale - Display predicted values in log scale
Sequence Highlight - Highlight motif based on Motif contribution or TSS contribution
Target - Choose among different prediction target files
Thank you for using TSS Analyzer. If you have any question or feedback, you can let us know at https://github.com/jzhoulab/puffin/discussions.